Use LogBert to do log anomaly detection
1. General Knowledge about Bert
Reference:
- Fine-tuning BERT for Text classification in Kaggle
- Natural Language Processing IN2361 by Prof. Dr. Georg Groh
BERT (Bidirectional Encoder Representations from Transformers)
Language modeling is a common method of pretraining on unlabeled text (self supervised learning). Most of the language models learned by iteratively predicting next word in a sequence auto regressively across enormous data sets of text like wikepedia. This can be left to right, right to left or bi-directional.
There are two strategies of applying pretrained language representations to downstream tasks:
- Feature based approach
- Fine tuning approach
The feauture based approach, such as ELMo uses task specific architectures that include the pretrained representations as additional features.
The fine tuning approach, such as OpenAI GPT, introduces minimal task specific parameters, and is trained on the downstream task by fine tuning all the pretrained parameters.
BERT model can be used for both the approaches. BERT reformulates the language modeling pretrained task of iteratively predicting the next word in sequence to instead incorporate bidirectional context and predict mask of intermediate tokens of the sequence and predict the mask token. BERT presented a new self supervised learning task for pretaining transformers in order to fine tune them for different tasks. They major difference between BERT and prior methods of pretraining transformer models is using the bidirectional context of language modeling. Most of the models either move left to right or right to left to predict next word in sequence, where BERT tries to learn intermediate tokens (by MASK), making the name Bidirectional Encoder.
BERT uses Masked language model and also use “Next sentence prediction” task.
BERT uses 3 embeddings to compute the input representations. They are token embeddings, segment embeddings and position embeddings.
BERT Transformer will preserve the length of the (dimention of the) input. The final output will take this vector and pass these to seperate tasks (classification, in this case).
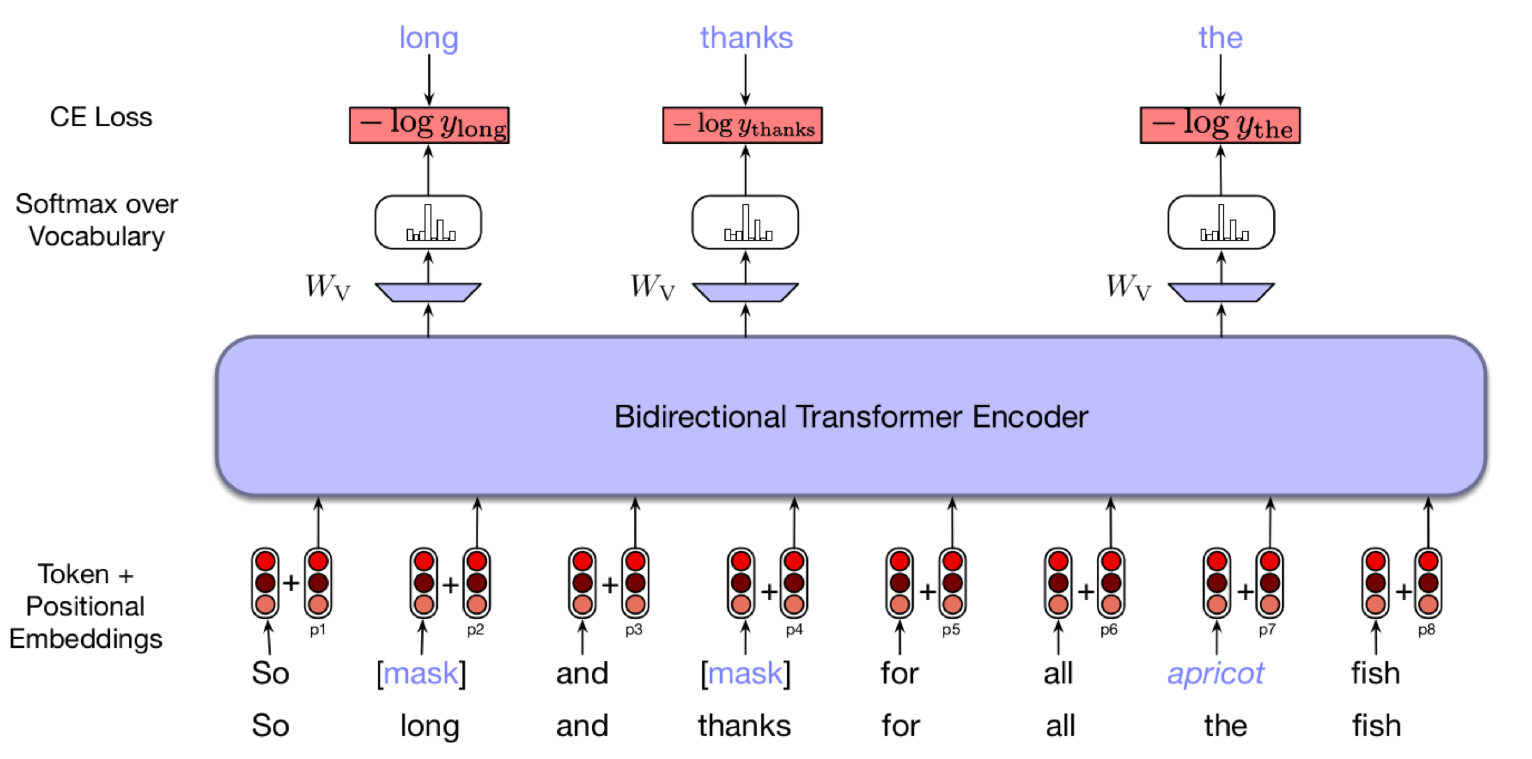
Bert Training Process with masked language model
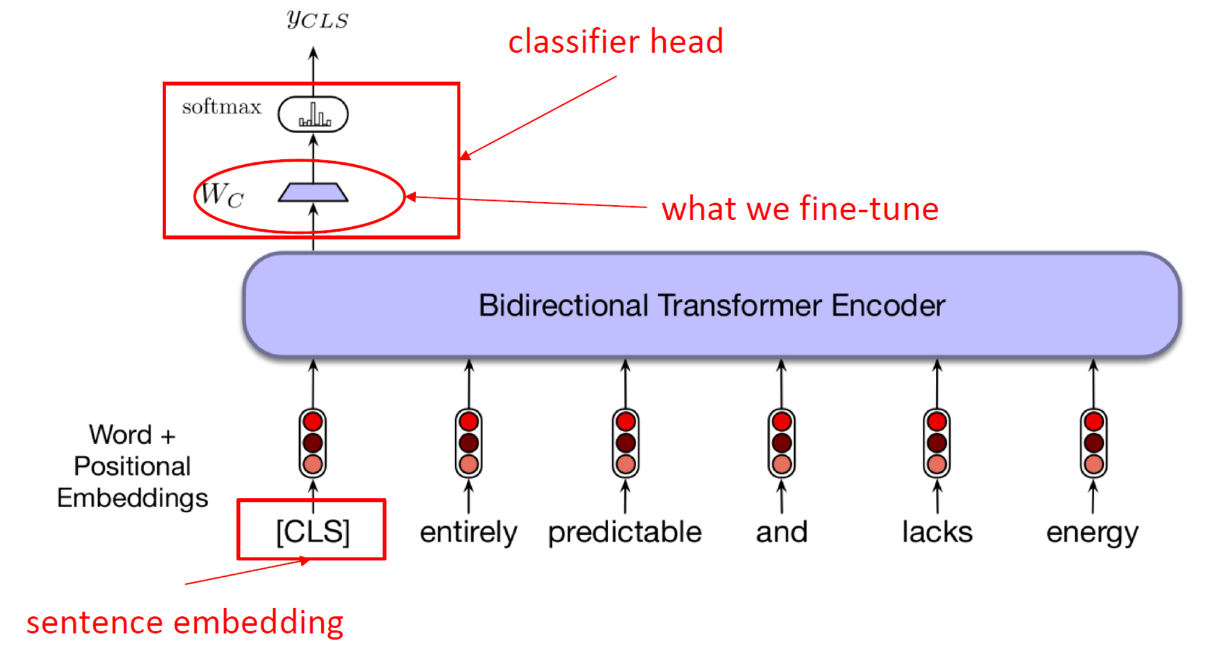
BERT for Classification [LogBert didn’t use this strategy]
BERT consists of stacked encoder layers. Just like the input of encoder of the transformer model, BERT model takes the sequence of numeric representation of the tokens as input. For classification tasks, we must prepend the special [CLS] token (classification token)to the beginning of every sentence.
Encoder block of transformer outputs a vector with same length as of input. First position of the vector, corresponding to the [CLS] token, can now be used as the input for a classifier.
2. How to use Masked Langauge Model to do Anomaly Detection [LogBert method]
[Guo, Haixuan, Shuhan Yuan, and Xintao Wu. “Logbert: Log anomaly detection via bert.” 2021 international joint conference on neural networks (IJCNN). IEEE, 2021.]
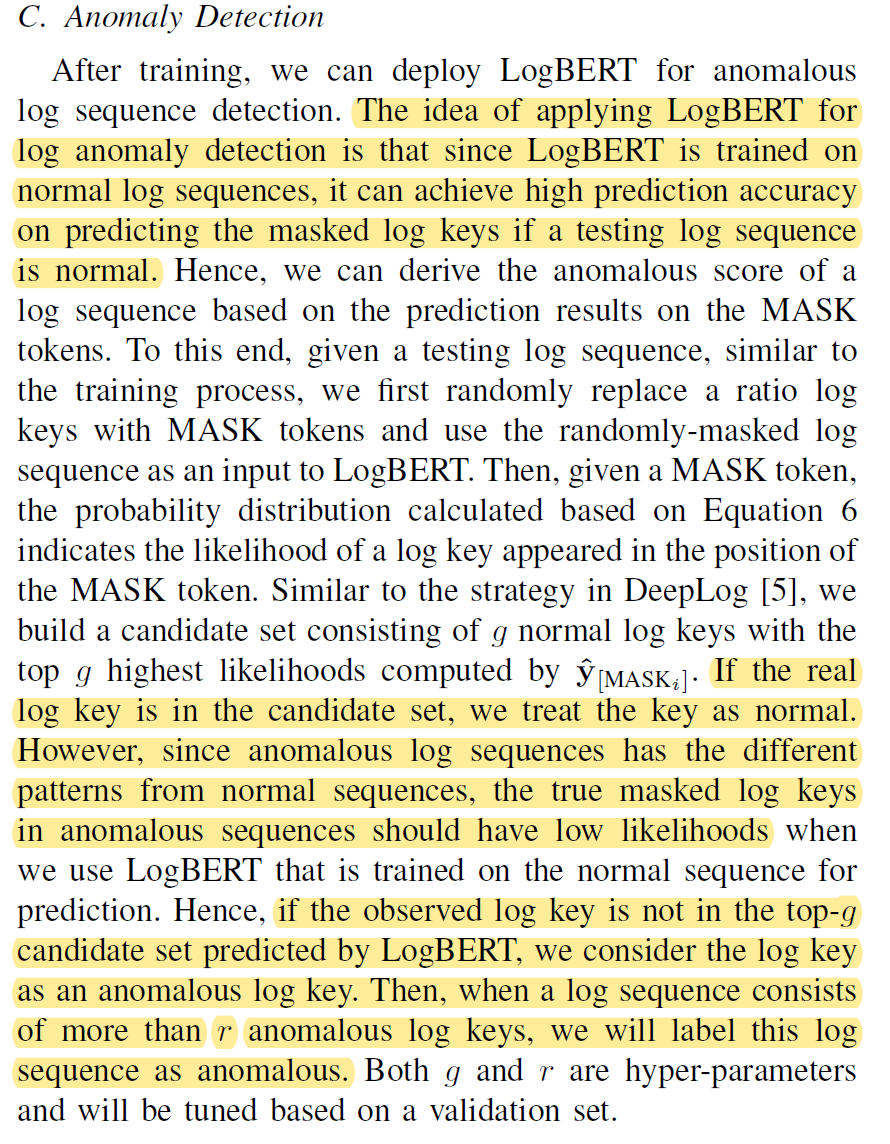
General Idea:
- Finetune the pretrained Bert model on the normal log lines by training on the masked language model.
- After this finetuning process, the model should predict the masked words in normal log lines very well. Since it didn’t see any abnormal logs before, it should predict the masked words in abnormal log lines very badly.
- From this point, we can seperate normal log lines and abnormal lines by looking at the performance of the model, which is reflected by the entropy loss value of prediction.
3. LogBert in LogAI - the whole pipeline
3.1 Load Opensearch indices, preprocessing and save as pandas “feather” format
the util function that load data in range:
def load_data_range(index_name, start, end, client=None):
if client == None:
client = OpenSearch(
hosts=['https://10.11.40.114:9200'],
use_ssl=True,
verify_certs=False,
http_auth=('admin', 'admin'),
timeout=60
)
max_index = client.indices.stats(index=index_name)['_all']['primaries']['docs']['count'] - 1
start = min(start, max_index)
end = min(end, max_index)
print("Loading data from index " + str(start) + " until index " + str(end) + " size is " + str(end - start + 1))
query = {
"from": start,
"size": end - start + 1,
"query": {
"match_all": {}
},
}
search_results = client.search(index=index_name, body=query)
# Extract and convert the documents to a Pandas DataFrame
documents = search_results["hits"]["hits"]
data = [doc['_source'] for doc in documents]
df = pd.DataFrame(data)
return df
init opensearch client, read the index in pieces and concat them together and save them:
import warnings
from collections.abc import MutableMapping
warnings.filterwarnings('ignore')
import pandas as pd
from opensearchpy import OpenSearch
import math
import pickle
import utils
client = OpenSearch(
hosts=['https://10.11.40.114:9200'],
use_ssl=True,
verify_certs=False,
http_auth=('admin', 'admin'),
timeout=60
)
index_name = "2023-09-05_system_v4"
size = client.indices.stats(index=index_name)['_all']['primaries']['docs']['count']
iteration_size = 500000
dataset_iterations = math.ceil((size / iteration_size))
print("df size: " + str(size) + " iterations: " + str(dataset_iterations))
df_save = pd.DataFrame()
for i in range(0, dataset_iterations):
print("Iteration " + str(i + 1) + " of " + str(dataset_iterations))
df = utils.load_data_range(index_name, (iteration_size * i), ((iteration_size * (i + 1)) - 1), client)
df_save = pd.concat([df_save, df])
df_save = df_save.reset_index()
df_save.to_feather('filename.feather')
some preprocessing on the raw opensearch data:
import pandas as pd
## read saved feather data
training_df1 = pd.read_feather('filename1.feather')
training_df2 = pd.read_feather('filename2.feather')
testing_df = pd.read_feather('filename3.feather')
## rename logline column from "desciption" to "logline", because logai uses constant## "logline" as the default column name of logline.
testing_df.rename(columns={'description':'logline'},inplace=True)
training_df1.rename(columns={'description':'logline'},inplace=True)
training_df2.rename(columns={'description':'logline'},inplace=True)
## set span_id
testing_df['span_id'] = testing_df['index']
training_df1['span_id'] = training_df1['index']
training_df2['span_id'] = training_df2['index']
## label anomalies, note that the training process is purely unsupervised and## doesn't require labels, but it seems necessary to assign labels column to the## dataframe. Also it helps visualize the result later if you want to see the## loglines that you are interested in (just lable those interesting log lines with 1)## In normal cases, just set the whole columns 0 values
training_df1["labels"] = 0
training_df2["labels"] = 0
testing_df["labels"] = 0
for index, row in testing_df.iterrows():
if "ABEND" in row['logline']:
testing_df.at[index, 'labels'] = 1
if "ERROR" in row['logline']:
testing_df.at[index, 'labels'] = 1
if "WARN" in row['logline']:
testing_df.at[index, 'labels'] = 1
## concat 2 training dataframe as one and reset index, span_id
training_df = pd.concat([training_df1,training_df2]).reset_index()
training_df['index'] = training_df.index
training_df['span_id'] = training_df['index']
training_df.drop(['level_0'],axis=1, inplace=True)
testing_df['index'] = testing_df.index
testing_df['span_id'] = testing_df['index']
## save them again
testing_df.to_pickle('testing_df.pkl')
training_df.to_pickle('training_df.pkl')
3.2 Custom preprocessor
read the saved pkl from the last step:
testing_df = pd.read_pickle('testing_df.pkl')
training_df = pd.read_pickle('training_df.pkl')
To encode special values (like IP, HEX value, INT value, etc.) into special tokens and preprocess , we need to override 2 methods from OpenSetPreprocessor, specify the preprocessor_config, and set special tokens in the tokenizer.
3.2.1 Override 2 methods from OpenSetPreprocessor
- _get_ids(self, logrecord) → pd.Series
- _get_labels(self, logrecord)
from logai.preprocess.openset_preprocessor import OpenSetPreprocessor
class SDMLOG_Preprocessor(OpenSetPreprocessor):
"""
Custom preprocessor for Open log dataset BGL.
"""
def __init__(self, config: PreprocessorConfig):
super().__init__(config)
def _get_ids(self, logrecord: LogRecordObject) -> pd.Series:
"""Get ids of loglines.
:param logrecord: logrecord object containing the BGL data.
:return: pd.Series object containing the ids of the loglines.
"""
ids = logrecord.span_id[constants.SPAN_ID]
return ids
def _get_labels(self, logrecord: LogRecordObject):
"""Get anomaly detection labels of loglines.
:param logrecord: logrecord object containing the BGL data.
:return:pd.Series object containing the labels of the loglines.
"""
return logrecord.labels[constants.LABELS]
preprocessor = SDMLOG_Preprocessor(config.preprocessor_config)
preprocessed_filepath = os.path.join(config.output_dir, 'filename.csv')
logrecord = preprocessor.clean_log(logrecord)
logrecord.save_to_csv(preprocessed_filepath)
3.2.2 specify preprocessor-related config
preprocessor_config:
custom_delimiters_regex:
[':', ',', '=', '\\t']
custom_replace_list: [
['(0x)[0-9a-zA-Z]+', ' HEX '],
['((?![A-Za-z]{8}|\\d{8})[A-Za-z\\d]{8})', ' ALPHANUM '],
['\\d+.\\d+.\\d+.\\d+', ' IP '],
['\\d+', ' INT ']
]
3.2.3 set special tokens in the tokenizer config, which should be the same list of “custom_replace_list” in the preprocessor_config
log_vectorizer_config:
algo_name: "logbert"
algo_param:
model_name: "bert-base-cased"
max_token_len: 120
custom_tokens: ["ALPHANUM", "IP", "HEX", "INT"]
output_dir: "outputdir"
tokenizer_dirname: "logbert_tokTenizer"
3.3 Split the training, validation and testing dataset
use sklearn.model_selection train_test_split:
here I split the logrecord_training into 99% training data, and 1% evaluation data, for the sake of adequate amount of training data and quick evaluation
from sklearn.model_selection import train_test_split
train_filepath = os.path.join(config.output_dir, 'train.csv')
dev_filepath = os.path.join(config.output_dir, 'dev.csv')
test_filepath = os.path.join(config.output_dir, 'test.csv')
train_ids, dev_ids, train_labels, dev_labels = train_test_split(
logrecord_training.span_id[constants.SPAN_ID],
logrecord_training.labels[constants.LABELS],
test_size=0.01,
shuffle=False,
stratify=None,
)
indices_train = list(
logrecord_training.span_id.loc[
logrecord_training.span_id[constants.SPAN_ID].isin(train_ids)
].index
)
indices_dev = list(
logrecord_training.span_id.loc[logrecord_training.span_id[constants.SPAN_ID].isin(dev_ids)].index
)
indices_test = list(
logrecord_testing.span_id.index
)
train_data = logrecord_training.select_by_index(indices_train)
dev_data = logrecord_training.select_by_index(indices_dev)
test_data = logrecord_testing.select_by_index(indices_test)
#train_data.save_to_csv(train_filepath)#dev_data.save_to_csv(dev_filepath)#test_data.save_to_csv(test_filepath)
print ('Train/Dev/Test Anomalous', len(train_data.labels[train_data.labels[constants.LABELS]==1]),
len(dev_data.labels[dev_data.labels[constants.LABELS]==1]),
len(test_data.labels[test_data.labels[constants.LABELS]==1]))
print ('Train/Dev/Test Normal', len(train_data.labels[train_data.labels[constants.LABELS]==0]),
len(dev_data.labels[dev_data.labels[constants.LABELS]==0]),
len(test_data.labels[test_data.labels[constants.LABELS]==0]))
3.4 Train Bert vectorizer
vectorizer = LogVectorizer(config.log_vectorizer_config)
vectorizer.fit(train_data)
train_features = vectorizer.transform(train_data)
dev_features = vectorizer.transform(dev_data)
test_features = vectorizer.transform(test_data)
3.5 Bert model training
related config:
nn_anomaly_detection_config:
algo_name: "logbert"
algo_params:
model_name: "bert-base-cased"
learning_rate: 0.00005
num_train_epochs: 1
per_device_train_batch_size: 32
max_token_len: 120
save_steps: 1000
eval_steps: 1000
tokenizer_dirpath: "bert-base-cased_tokenizer"
output_dir: "outputdir"
pretrain_from_scratch:False
anomaly_detector = NNAnomalyDetector(config=config.nn_anomaly_detection_config)
anomaly_detector.fit(train_features, dev_features)
3.6 Bert model testing
related config:
nn_anomaly_detection_config:
algo_name: "logbert"
algo_params:
model_name: "bert-base-cased"
per_device_eval_batch_size: 32
eval_accumulation_steps: 284
mask_ngram: 8
tokenizer_dirpath: "bert-base-cased_tokenizer"
output_dir: "outputdir"
num_eval_shards: 100
anomaly_detector = NNAnomalyDetector(config=config.nn_anomaly_detection_config)
predict_results = anomaly_detector.predict(test_features_piece)
print (predict_results)
3.7 Output Intepretation
take partitioned logs as example, here is the data flow:
- prediction.feather
- 406105x19cols
- 349 anomalies
↓ Partitioner based on span_id, here span_id = seconds
- 323 lines + labels(1 if merged lines contain anomalies otherwise 0)
↓ Train dev test split
-
splited result
indices_train/dev/test: 181 20 122
Train/Dev/Test Anomalous: 0 0 71
Train/Dev/Test Normal: 181 20 51
↓ Turn pandas df into LogRecordObject
- LogRecordObject test_data, keys: [Timestamp, attributes, span_id, body, labels, _index]
↓ LogVectorizer
- test_features, features: [‘labels’, ‘input_ids’, ‘token_type_ids’, ‘attention_mask’], num_rows: 122
- for one row:
label: [1] input_ids size: 120 value: [[2, 1378, 584, 5, 1278, 855, 3, 1548, 855, 584, 5, 1278, 1287, 11, 11, 1729, 23, 50, 11, 401, 11, 1565, 11, 1287, 51, 139, 3, 1160, 855, 968, 821, 2087, 3, 576, 864, 454, 2075, 10, 2123, 17, 11, 182, 18, 3, 2082, 2107, 9, 2035, 312, 2126, 3, 1685, 1160, 1339, 3, 1685, 1339, 2119, 3, 2046, 1160, 2101, 3, 1965, 576, 2056, 10, 2027, 2055, 1010, 1964, 1691, 3, 1160, 855, 454, 908, 3, 42, 563, 406, 94, 75, 462, 216, 454, 1419, 206, 1041, 95, 85, 3, 182, 32, 549, 1650, 71, 9, 185, 454, 672, 264, 1693, 1346, 10, 1334, 139, 10, 139, 5, 1333, 139, 5, 393, 5, 408, 428, 1315, 11, 3]] token_type_ids size: 120 value: [[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]] attention_mask size: 120 value: [[1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1]]
- for one row:
↓ NNAnomalyDetector.predict _generate_masked_input
-
mlm_dataset_test [masked language model]
Dataset({ features: [‘labels’, ‘input_ids’, ‘token_type_ids’, ‘attention_mask’, ‘indices’], num_rows: 1348 })
- for one row:
Labels: [[-100, 1378, 584, -100, 1278, 855, -100, 1548, 855, 584, -100, 1278, 1287, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100, -100]] Input_ids size: 120 value: [[2, 4, 4, 5, 4, 4, 3, 4, 4, 4, 5, 4, 4, 11, 11, 1729, 23, 50, 11, 401, 11, 1565, 11, 1287, 51, 139, 3, 1160, 855, 968, 821, 2087, 3, 576, 864, 454, 2075, 10, 2123, 17, 11, 182, 18, 3, 2082, 2107, 9, 2035, 312, 2126, 3, 1685, 1160, 1339, 3, 1685, 1339, 2119, 3, 2046, 1160, 2101, 3, 1965, 576, 2056, 10, 2027, 2055, 1010, 1964, 1691, 3, 1160, 855, 454, 908, 3, 42, 563, 406, 94, 75, 462, 216, 454, 1419, 206, 1041, 95, 85, 3, 182, 32, 549, 1650, 71, 9, 185, 454, 672, 264, 1693, 1346, 10, 1334, 139, 10, 139, 5, 1333, 139, 5, 393, 5, 408, 428, 1315, 11, 3]] token_type_ids size: 120 value: [[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]] attention_mask size: 120 value: [[1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1]] - what does _generate_masked_input do?
- add masks on raw dataset:
examples1 = {'input_ids' : [[10,5,7,-1,2]], 'attention_mask' : [[1,1,1,1,1]], 'token_type_ids' : [[0,0,0,0,0]]} indices = [0] output = generate_masked_input(examples1, indices) output {'input_ids': array([[ 4, 5, 7, -1, 2], [10, 4, 7, -1, 2], [10, 5, 4, -1, 2], [10, 5, 7, -1, 4]], dtype=int64), 'attention_mask': array([[1, 1, 1, 1, 1], [1, 1, 1, 1, 1], [1, 1, 1, 1, 1], [1, 1, 1, 1, 1]]), 'token_type_ids': array([[0, 0, 0, 0, 0], [0, 0, 0, 0, 0], [0, 0, 0, 0, 0], [0, 0, 0, 0, 0]]), 'labels': array([[ 10, -100, -100, -100, -100], [-100, 5, -100, -100, -100], [-100, -100, 7, -100, -100], [-100, -100, -100, -100, 2]]), 'indices': array([0, 0, 0, 0], dtype=int64)}
- for one row:
↓ NNAnomalyDetector.predict predict
-
test_results
Test_results [‘predictions’, ‘label_ids’, ‘metrics’]
- Metrics: [‘test_loss’,’test_runtime’,’test_samples_per_second’,’test_steps_per_second]
-
One shard: [to avoid OOM, the whole mlm_dataset_test is split into 10 shards, and do prediction on each shard]
Predictions.shape: (135,120,2220) Label_ids.shape: (135,120) 135*10-2 = 1348
-
All result shape: (1348,120,2220)
2220 is the volabulary size
120 is the seq length
1348 is the # of seqs
-
intepretation
filtered_result = predict_result[5, [10,14,20,25], :] *# filtered_result has a shape of (4,2220)# for each 2220 vector, it shows the prob distribution of words behind this mask# you can get the output probability of real token_ids:* prob1 = predict_result[5,10,4] prob2 = predict_result[5,14,6] prob3 = predict_result[5,20,8] prob4 = predict_result[5,25,11] *# calculate cross entropy loss: -p*log(p)* entropy_i = -prob_i * log(prob_i) *# get a list of losses* entropies = [entropy1, entropy2, entropy3, entropy4, ...] *# calculate top 6 losses and take the mean of them as anomaly score:* anomaly score = mean(top6(entropies)) *# after this process, we can get a anomaly scores of shape (1348,)*The Bert output has the same dimension of input, in this case the input is the mlm_dataset_test (1348,120), embed each token_id into 2220 dictionary vector, then it’s (1348,120, 2220), so the output is the same dimension of (1348,120,2220)
for each log lines, we only need to calculate the loss function on the masked words, which were specified by a special token type [mask].
for example, let’s assume in the masked logline 5, the index [10,14,20,25] are 4 words positions with [mask], and their token_ids are [4, 6, 8, 11]. We fetch those data out:
↓ anomaly_results.groupby('indices').mean()
- aggregate the anomaly score of the same index, calculate the mean of them as the final anomaly score of this log line.
3.8 The code modification for the use of GPU acceleration and no partitioning
-
in logai\logai\algorithms\nn_model\logbert[train.py](http://train.py/):
intend to use GPU if the environment has, not sure if this change is necessary, because I had experience that forgot to change train.py but still trained with GPU.
import torch
def _initialize_trainer(self, model, train_dataset, dev_dataset):
"""initializing huggingface trainer object for logbert"""
training_args = TrainingArguments(
self.model_dirpath,
evaluation_strategy=self.config.evaluation_strategy,
num_train_epochs=self.config.num_train_epochs,
learning_rate=self.config.learning_rate,
logging_steps=self.config.logging_steps,
per_device_train_batch_size=self.config.per_device_train_batch_size,
per_device_eval_batch_size=self.config.per_device_eval_batch_size,
weight_decay=self.config.weight_decay,
save_steps=self.config.save_steps,
eval_steps=self.config.eval_steps,
resume_from_checkpoint=self.config.resume_from_checkpoint,
)
data_collator = DataCollatorForLanguageModeling(
tokenizer=self.tokenizer,
mlm_probability=self.config.mlm_probability,
pad_to_multiple_of=self.config.max_token_len,
)
## modified by huiyu
if torch.cuda.is_available():
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
self.trainer = Trainer(
model=model.to(device),
args=training_args,
train_dataset=train_dataset,
eval_dataset=dev_dataset,
data_collator=data_collator,
)
else:
self.trainer = Trainer(
model=model,
args=training_args,
train_dataset=train_dataset,
eval_dataset=dev_dataset,
data_collator=data_collator,
)
- in logai\logai\algorithms\nn_model\logbert[predict.py](http://predict.py/):
- I partitioned the log lines in the beginning because in the condition of no partitioning, during the execution of _generate_masked_input, it prompted an error “num_sections must be bigger than 0”, because there were cases that the whole log line contains purely so called special_token_ids.
- Added code ensures that lines with pure special tokens should return all labels with -100 so that they will not be taken into account when calculating the loss function in the later phase; in simple words, those lines will be neglected.
def _generate_masked_input(self, examples, indices):
input_ids = examples["input_ids"][0]
attention_masks = examples["attention_mask"][0]
token_type_ids = examples["token_type_ids"][0]
index = indices[0]
input_ids = np.array(input_ids)
sliding_window_diag = np.eye(input_ids.shape[0])
if sliding_window_diag.shape[0] == 0:
return examples
mask = 1 - np.isin(input_ids, self.special_token_ids).astype(np.int64)
sliding_window_diag = sliding_window_diag * mask
sliding_window_diag = sliding_window_diag[
~np.all(sliding_window_diag == 0, axis=1)
]
## modified by huiyu
if sliding_window_diag.shape[0] == 0:
output = {}
output["input_ids"] = np.array(examples["input_ids"])
output["attention_mask"] = np.array(examples["attention_mask"])
output["token_type_ids"] = np.array(examples['token_type_ids'])
output["labels"] = -100 * np.ones(shape=(output["input_ids"].shape[0],output["input_ids"].shape[1]))
output["indices"] = np.array([index]).astype(
np.int64
)
return output
##
num_sections = int(sliding_window_diag.shape[0] / self.config.mask_ngram)
if num_sections <= 0:
num_sections = sliding_window_diag.shape[0]
sliding_window_diag = np.array_split(sliding_window_diag, num_sections, axis=0)
diag = np.array([np.sum(di, axis=0) for di in sliding_window_diag])
input_rpt = np.tile(input_ids, (diag.shape[0], 1))
labels = np.copy(input_rpt)
input_ids_masked = (input_rpt * (1 - diag) + diag * self.mask_id).astype(
np.int64
)
attention_masks = np.tile(
np.array(attention_masks), (input_ids_masked.shape[0], 1)
)
token_type_ids = np.tile(
np.array(token_type_ids), (input_ids_masked.shape[0], 1)
)
labels[
input_ids_masked != self.mask_id
] = -100 # Need masked LM loss only for tokens with mask_id
examples = {}
examples["input_ids"] = input_ids_masked
examples["attention_mask"] = attention_masks
examples["token_type_ids"] = token_type_ids
examples["labels"] = labels
examples["indices"] = np.array([index] * input_ids_masked.shape[0]).astype(
np.int64
)
return examples
-
in logai\logai\algorithms\vectorization_algo\logbert.py:
This is one crutial change to the logai package. In the following _clean_dataset() method, log lines with pure special tokens will be excluded. The problem is that we don’t know the index of included lines when this function returns.
- To get the original indices, we need to return “old indices” from cleandataset() and return again in transform();
-
Also change the receiving params in fit(): cleaned_logrecord, _ = self._clean_dataset(logrecord)
## modified by Huiyu ## return indices from _clean_dataset, and return them in transform() def _clean_dataset(self, logrecord: LogRecordObject): special_tokens = self._get_all_special_tokens() loglines = logrecord.body[constants.LOGLINE_NAME] loglines_removed_special_tokens = loglines.apply( lambda x: " ".join(set(x.split(" ")) - set(special_tokens)).strip() ) indices = list(logrecord.body.loc[loglines_removed_special_tokens != ""].index) logrecord = logrecord.select_by_index(indices, inplace=True) return logrecord, indices def transform(self, logrecord: LogRecordObject): """Transform method for running vectorizer over logrecord object. :param logrecord: A log record object containing the dataset to be vectorized. :return: HuggingFace dataset object. """ cleaned_logrecord, old_indices = self._clean_dataset(logrecord) dataset = self._get_hf_dataset(cleaned_logrecord) tokenized_dataset = dataset.map( self._tokenize_function, batched=True, num_proc=self.config.num_proc, remove_columns=[constants.LOGLINE_NAME], ) return tokenized_dataset, old_indices ## def fit(self, logrecord: LogRecordObject): """Fit method for training vectorizer for logbert. :param logrecord: A log record object containing the training dataset over which vectorizer is trained. """ if os.listdir(self.config.tokenizer_dirpath): return #also change here cleaned_logrecord, _ = self._clean_dataset(logrecord) dataset = self._get_hf_dataset(cleaned_logrecord) def batch_iterator(): for i in range(0, len(dataset), self.config.train_batch_size): yield dataset[i : i + self.config.train_batch_size][ constants.LOGLINE_NAME ] trainer = trainers.WordPieceTrainer( vocab_size=self.config.max_vocab_size, special_tokens=self.special_tokens ) self.tokenizer.train_from_iterator(batch_iterator(), trainer=trainer) cls_token_id = self.tokenizer.token_to_id("[CLS]") sep_token_id = self.tokenizer.token_to_id("[SEP]") self.tokenizer.post_processor = processors.TemplateProcessing( single=f"[CLS]:0 $A:0 [SEP]:0", pair=f"[CLS]:0 $A:0 [SEP]:0 $B:1 [SEP]:1", special_tokens=[ ("[CLS]", cls_token_id), ("[SEP]", sep_token_id), ], ) self.tokenizer.decoder = decoders.WordPiece(prefix="##") new_tokenizer = BertTokenizerFast(tokenizer_object=self.tokenizer) new_tokenizer.save_pretrained(self.config.tokenizer_dirpath) self.tokenizer = new_tokenizer